Framework to Design mRNA Sequences to Control Co-Translational Folding
ID# 2015-4330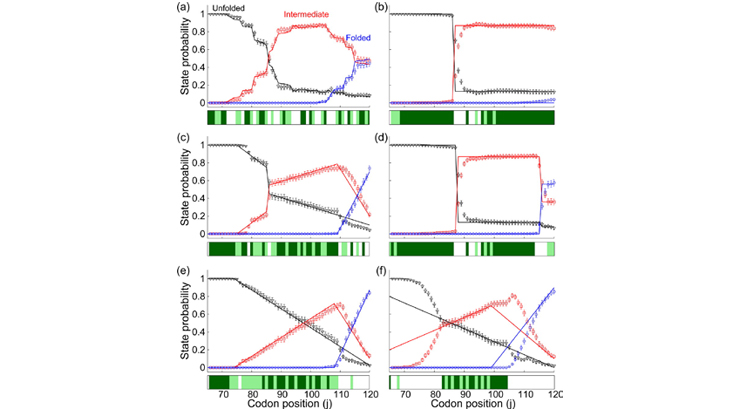
Technology Summary
This invention allows control of the co-translational folding of a protein through synonymous codon mutations. It uses a Monte Carlo master equation-based framework to generate a large number of degenerate sequences whose translation produces the desired folding behavior of a nascent protein. Traditional codon optimization methods enhance the produc¬tion of proteins in heterologous gene expression, but proteins produced from these optimized mRNA sequences misfold/aggregate. This invention solves this issue, enhancing quality of heterologous protein production by maximizing proper folding and structure. The invention allows control of the folding of newly created proteins within cells by altering the mRNA sequence without changing the amino acid. This method uses an algorithm with repeated random sampling to generate sequences with similar mRNA coding.
Application & Market Utility
High-quality proteins are required for commercial/academic applications. This method provides a way to rationally design mRNA sequences based on the folding kinetics of nascent protein. It predicts its own success or failure through mathematical modeling without actually carrying out an experiment, thus reducing costs and time. Applications include: Biotechnology companies that supply high-quality proteins to academic labs and other industries. Pharmaceutical companies that produce functional proteins to design monoclo¬nal antibodies or for therapeutic purposes.
Next Steps
Seeking research collaboration and licensing opportunities.

