Low Sequence Bias Single-Stranded DNA Ligation
ID# 2012-4006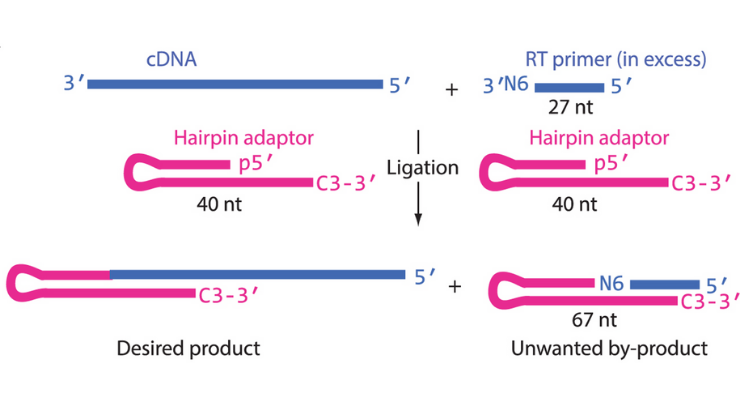
Technology Summary
Intermolecular single-stranded DNA ligation is important for biotechnical applications like Ligation-Mediated PCR and cDNA library construction. These require a fixed sequence DNA oligonucleotide to ligate to an unknown 3’-end of a cDNA. The subject invention hybridizes the incoming acceptor DNA oligonucleotide to a hairpin DNA enzymatically or chemically. The room temperature process is fast (<2 hrs), high yielding, reproducible, carries low sequence bias, and integrates seamlessly with downstream protocols in comparison to Circligase I or T4 RNA ligase 1. The invention’s results are consistent with Mfold computer modeling predictions of the secondary structure of the ligated DNA product. For LMPCR type experiments, nucleotide preference ligation tests on four 24mer acceptors having each a different base at its 3’ end resulted in average yields above ninety percent.
Application & Market Utility
CircLigase and T4 suffer from slow kinetics, poor yield and severe sequence bias. Unaddressed sequence bias leads to misinterpretation of gene expression levels. This invention’s ligation method does not require a statistical correction factor for nucleotide bias. This method also allows the decision for NGS platform to be made at the last step of a protocol, allowing cross-platform validation on the same or different cDNA library samples. The resulting high throughput data will benefit from the greatly reduced sequence bias.
Next Steps
Seeking research collaboration and licensing opportunities.

