RNA-Guided Multiplex Genome Editing via the tRNA Processing System
ID# 2014-4242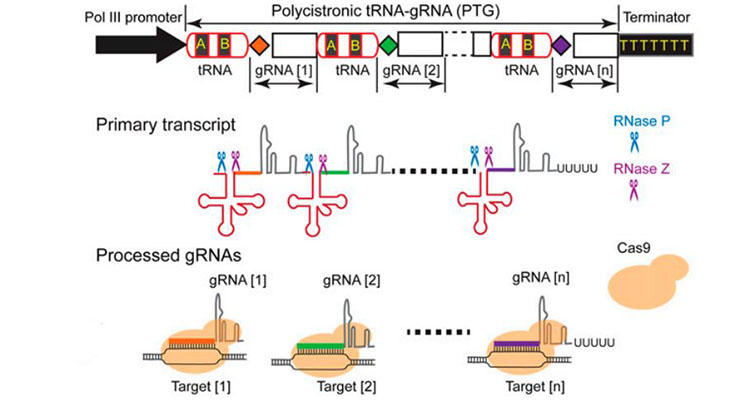
Technology Summary
The technology provides a method for ef?cient production of numerous gRNAs in vivo from a synthetic polycistronic gene via an endogenous RNA processing system. The technology represents an improved method that facilitates robust genome editing at multiple loci. Methods could be used to improve many RNA-based multiplex technologies, including but not limited to CRISPR/Cas9-mediated genome editing and modifications, epigenome editing, RNA targeting, transcriptional modulation, and artificial microRNA mediated gene silencing. The disclosed methods may additionally be used to insert exogenous DNA sequences into a predetermined location and target DNA sequences for knock-out or mutation via deletion. Disclosed methods and compositions have a broad range application for small RNA expression and genome engineering, additionally including cell culture and animal models.
Application & Market Utility
Highly precise and efficient cleavage for equal molar production of multiple gRNAs from a single transcript
No obligation to start with a specific nucleotide for U3 or U6 promoters
tRNA acts as an internal promoter to significantly incrase gRNA expression (~10 fold increase over U3 promoter)
Option to use a Pol II promoter and single transcriptional unit for efficient expression of gRNA/Cas in a spatial/temporal manner
Small insert size to facilitate viral delivery
Broad Application in Plants (exclusive licensee has sublicensing right) animals and microbes
Next Steps
Seeking research collaboration and licensing opportunities.

