IDEAS+: An Integrative and Discriminative Epigenome Annotation System
ID# 2017-4608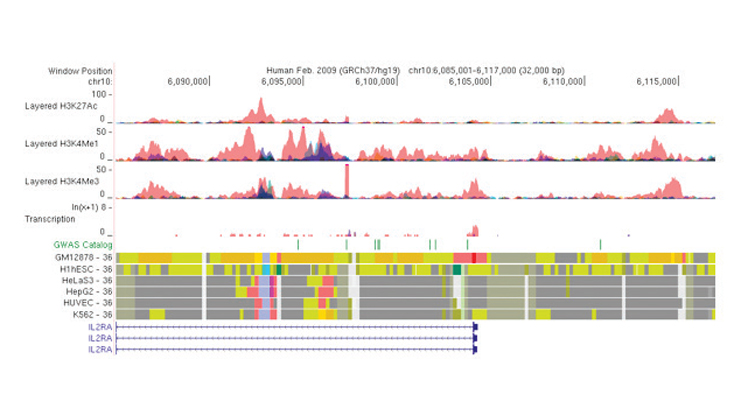
Technology Summary
The technology uses large-scale computational models to analyze thousands of functional data sets in hundreds of human cell types, tissues, and cancer cell lines to identify novel functions in the human genome. The predictions are then combined with human disease mutations identified in genetic studies to pinpoint disease causal variants and the genes that they may impact on. The technology can potentially much better predict functional mutations for complex disease, which currently is the bottleneck to identify the correct targets for treatment. The technology also enables detection of the most relevant cell types to the disease, in which experimental validation can be carried out to assess the computational predictions and then narrow down the set of drug targets.
Application & Market Utility
Inventors have used the technology to select functional mutations from a list of 4000 candidates and the most likely cell types in which they may be functional for complex diseases such as inflammatory bowel disease (IBD). It is a substantial breakthrough over existing pipelines for detecting genomic targets in disease treatment. The application of the technology can be substantial in drug discovery for pharmaceutical companies, and more broadly for communities in disease genetics and gene regulation.
Next Steps
Seeking research collaboration and licensing opportunities.

